%%html
<script src="https://bits.csb.pitt.edu/preamble.js"></script>
Central Dogma¶
DNA Sequence $\rightarrow$ RNA $\rightarrow$ Protein $\rightarrow$ Structure (Dynamics) $\rightarrow$ Function
import py3Dmol
Molecular Representations¶
Cartoons trace molecule and show key features.
v = py3Dmol.view('4c64',style='cartoon'); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
DNA Structure¶
Where is major/minor groove?
v = py3Dmol.view('4c64',style='cartoon'); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
Molecular Representations¶
Spheres (space-filling) highlights individual atoms (usually color coded by element).
v = py3Dmol.view('4c64',style='sphere'); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
Molecular Representations¶
Sticks (licorice) highlights bonds. Often used for small molecules.
v = py3Dmol.view('4c64',style='stick'); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
Note that sticks and cartoon won't show nonbonded atoms...
v = py3Dmol.view('4c64',style='stick')
v.setStyle({'bonds':0},{'sphere':{'radius':0.5}}); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
%%html
<div id="whatdots" style="width: 500px"></div>
<script>
var divid = '#whatdots';
jQuery(divid).asker({
id: divid,
question: "What are the red dots?",
answers: ["Water","Ions","Cofactors","Phosphates"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
Molecular Representations¶
Surfaces show the overall shape.
v = py3Dmol.view('4c64',style='stick',height=450)
v.addSurface().show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
Molecular Surfaces¶
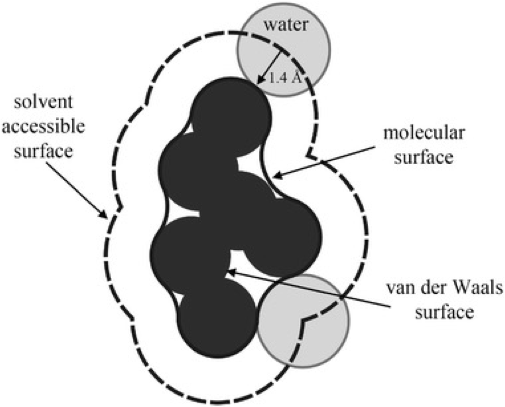
From Introduction to Proteins: Structure, Function, and Motion. Amit Kessel & Nir Ben-Tal
Molecular Surfaces¶
v = py3Dmol.view('4c64',width=770,viewergrid=(1,3))
v.addSurface('VDW',viewer=(0,0)); v.addSurface('MS',viewer=(0,1)); v.addSurface('SAS',viewer=(0,2)); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
RNA Can Have Interesting Structure¶
S-adenosylmethionine (SAM) / S-adenosylhomocysteine (SAH) riboswitch - regulates transcription of SAM-biosynthetic enzymes.
v = py3Dmol.view(query='6HAG',style='cartoon',height=450)
v.setStyle({'resn':'SAH'},'sphere').show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
RNA Can Have Interesting Structure¶
Transfer RNA
py3Dmol.view(query='4tna',style='stick').show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
...But We're Just Going To Focus on Proteins¶
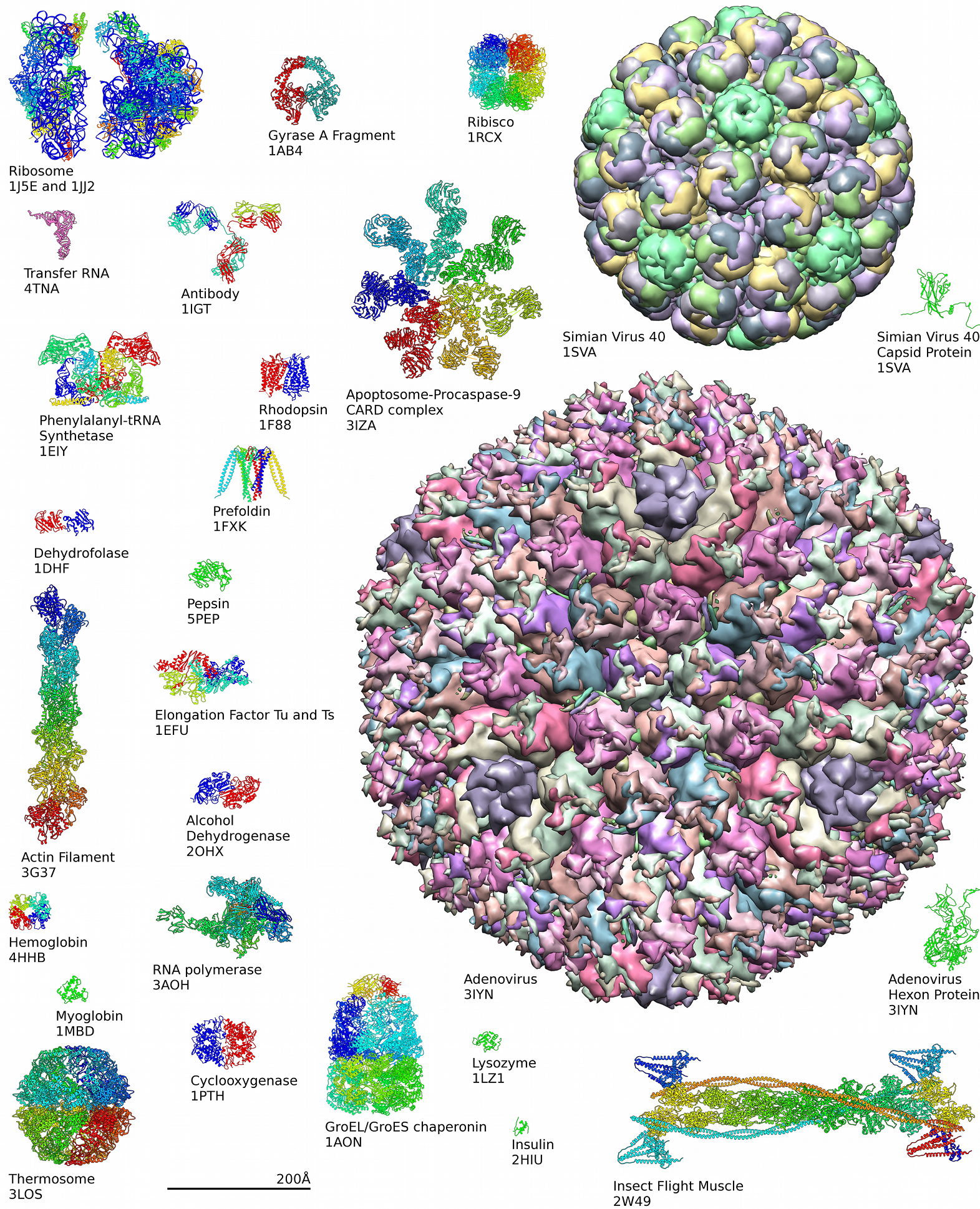
Proteins are important¶
- About half of dry mass of a cell
- Perform most of the cell's functions: transcription, signaling, catalysis, transport, molecular recognition, mechanical support, motion...
v = py3Dmol.view(query='3WTG',style={'cartoon':{'colorscheme':'chain'}},height=400)
v.setStyle({'resn':'HEM'},'stick').show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
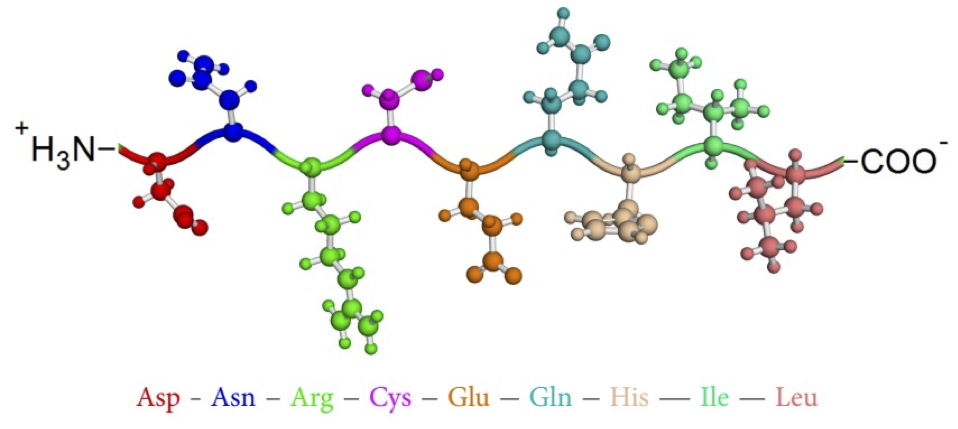
Proteins are polymers (chains) of amino acids¶
GATTACAGATTACAGATTACA $\rightarrow$ (N-terminal) DYRLQIT (C-terminal)
After forming the peptide bond, amino acids are called residues.
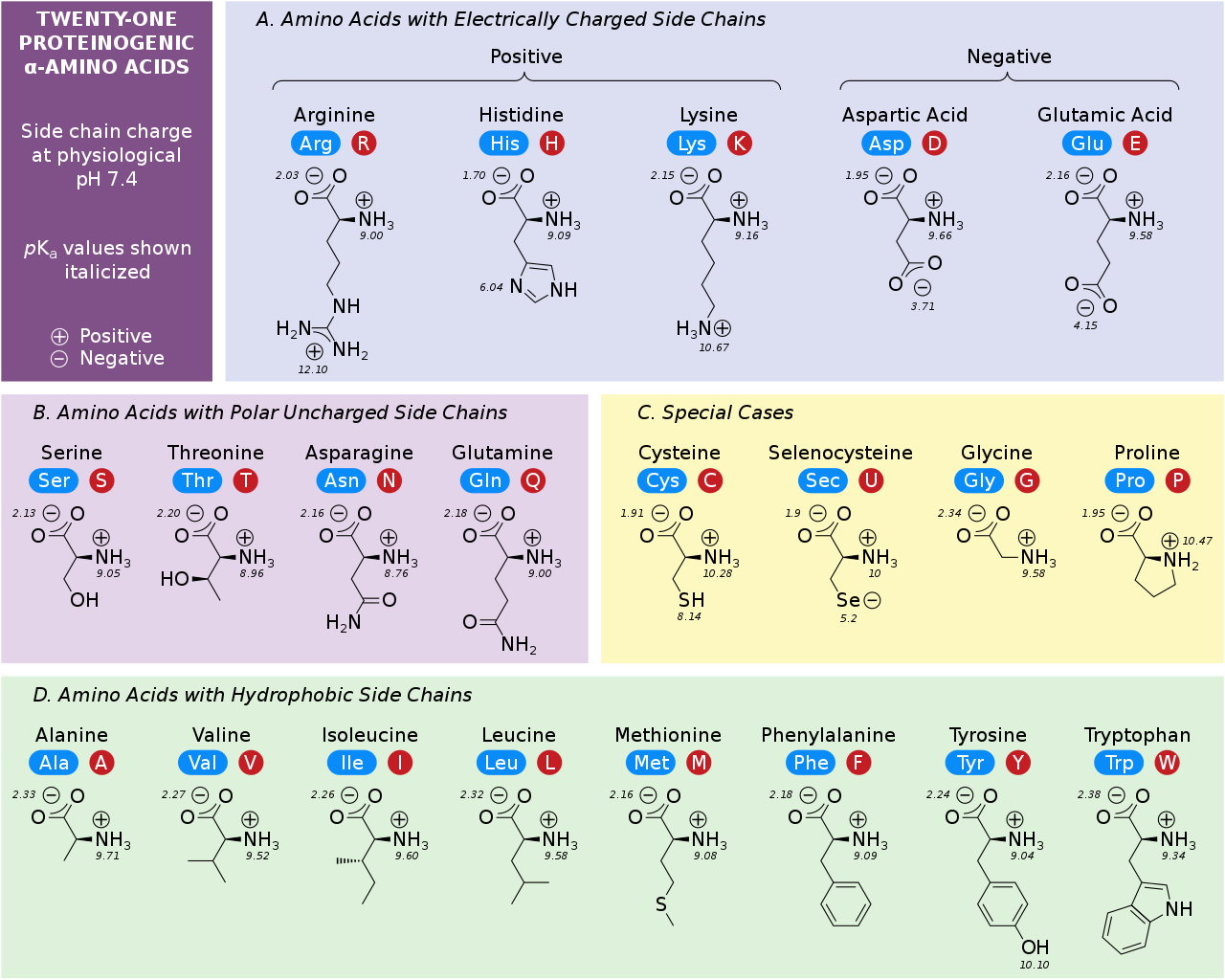
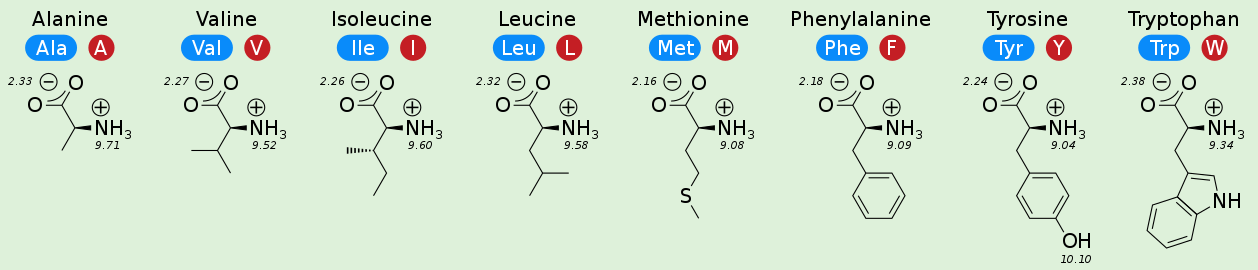
You need to know the 20 amino acids¶
v = py3Dmol.view(query='cid:5950',style={'stick':{},'sphere':{'radius':0.5}})
v.addLabel('C⍺',{'backgroundOpacity':.8},{'index':3});v.addLabel('N-terminal',{'backgroundOpacity':.8},{'index':10}); v.addLabel('C-terminal',{'backgroundOpacity':.8},{'index':5}); v.addLabel('Side-chain',{'backgroundOpacity':.8},{'index':8}); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
%%html
<div id="whataa" style="width: 500px"></div>
<script>
var divid = '#whataa';
jQuery(divid).asker({
id: divid,
question: "What is the previous amino acid?",
answers: ["alanine","asparigine",'glycine',"None of the above"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
pKa¶
Inverse measure of acid strength (lower number = stronger acid; opposite of Ka)
$$pK_a = -\log(K_a)$$
$$K_a = \frac{[R^-][H^+]}{[RH]}$$
$$pH = -\log([H^+]) = pK_a + \log\frac{[R^-]}{[RH]}$$
pKa¶
 $$pH = pK_a + \log\frac{[R^-]}{[RH]}$$
$$pH = pK_a + \log\frac{[R^-]}{[RH]}$$
- $pH > pK_a \rightarrow [R^-] > [RH]$ acid mostly deprotonated (hydrogen isn't there)
- $pH < pK_a \rightarrow [R^-] < [RH]$ acid mostly protonated (hydrogen is there)
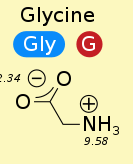
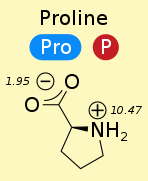
$pK_a$ of carboxyl group is ~2
$pK_a$ of amino group is ~9
%%html
<div id="pkaq" style="width: 500px"></div>
<script>
var divid = "#pkaq";
jQuery(divid).asker({
id: divid,
question: "At neutral pH, what is the pronation of the backbone amino and carboxyl groups?",
answers: ["N-,C-","N+,C-",'N-,C+',"N+,C+"],
extra: ["both deprotonated","N protonated, C deprotonated","N deprotonated, C protonated","both protonated"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
Side Chains Make Different Molecular Interactions¶
These molecular interactions determine the fold (shape) of the protein and contribute to its function.
- Charge - Charge
- Hydrogen bonding
- Aromaticity
- Hydrophobicity
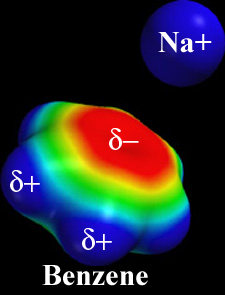
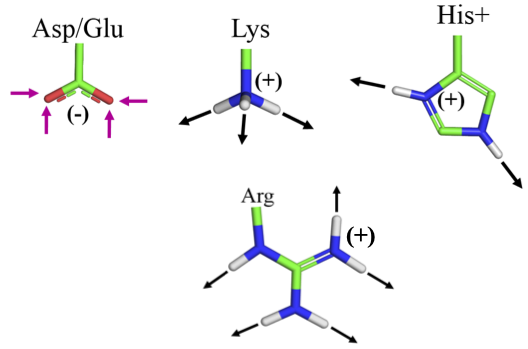
Charge - Charge¶
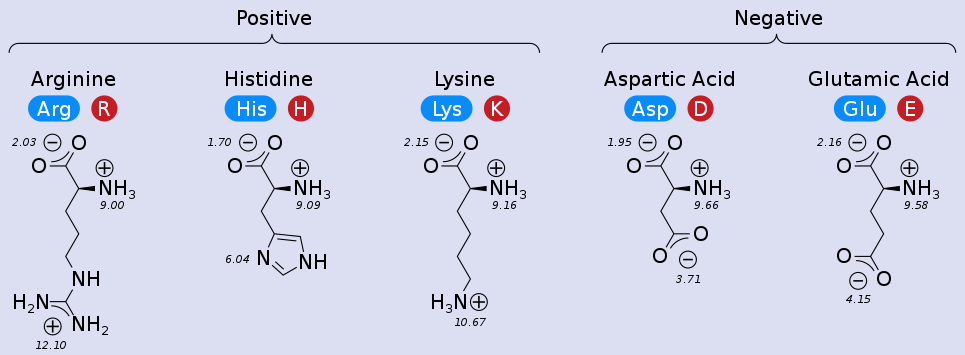
Charge - Charge¶
Coulomb's Law: the electrostatic force between two point charges is directly proportional to the product of the magnitudes of charges and inversely proportional to the square of the distance between them
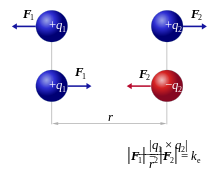
Note that the strength of the force depends on the environment: water will shield charges and lessen the force (more later).
These ionic interactions are called salt bridges.
Example: Lamin A¶
Structural protein of nuclear envelope. Phenotype of R527L is Mandibuloacral dysplasia (premature ageing)
v = py3Dmol.view(query='1ifr',style='cartoon'); sel = {'resi':[527,537]}; v.addStyle(sel,'stick'); v.zoomTo(sel); v.addResLabels(sel); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
%%html
<div id="saltbridge" style="width: 500px"></div>
<script>
var divid = "#saltbridge";
jQuery(divid).asker({
id: divid,
question: "Which pair of amino acids can NOT form a salt bridge?",
answers: ["R-D","L-D","K-D","R-E"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
Polar Groups¶

Polarization occurs when one of the atoms in a bond withdraws electrons towards (electronegativity) it resulting in partial charges on the atoms.
Polar molecules/groups have polar bonds.
Nonpolar molecules/groups do not have meaningfully polarized bonds (e.g., carbon-carbon).
Polar Hydrogens for Hydrogen Bonds¶
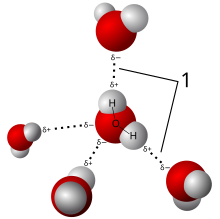
An electronegative atom (O or N) "shares" a hydrogen with another electronegative atom.
Strength depends on participating atoms, bond geometry (angle and distance), and environment.
Can be reasonably well approximated as purely electrostatic (dipole-dipole), but reality is more complicated.
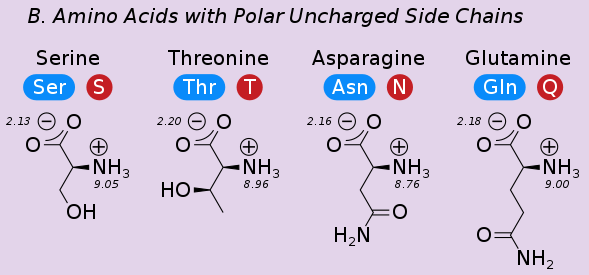
Hydrogen bonds are ubiquitous and involve the side-chains of polar (charged and uncharged) amino acids as well as the backbone of all amino acids.
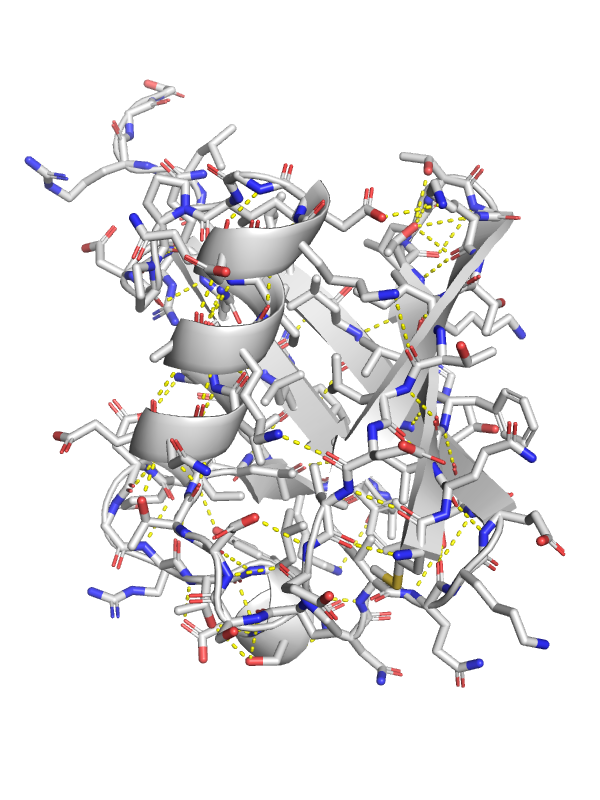
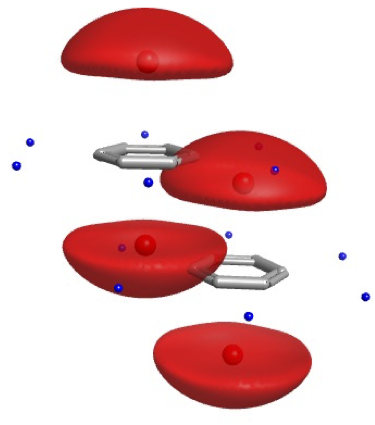
Hydrogen Bonds are Directional¶
From. Introduction to Proteins: Structure, Function, and Motion. Amit Kessel & Nir Ben-Tal
v = py3Dmol.view(query='6qtl',style='cartoon'); v.addStyle({'resi':[34,104]},'stick'); v.addStyle({'resi':[201]},{'stick':{'colorscheme':'greenCarbon'}}); v.zoomTo({'chain':'D','resi':201}); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
v = py3Dmol.view(query='2vab',style='cartoon'); sel = {'or':[{'chain':'A','resi':[66,167,170]},{'chain':'P','resi':1}]}; v.addStyle(sel,'stick'); v.zoomTo(sel); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
Hydrophobicity¶

The hydrophobic effect is not a force.
Nonpolar groups don't form favorable interactions with water.
These guys do not want to be solvent exposed and tend to pack in the hydrophobic core of the protein.
hydro = { 'prop': "resn", 'map': { 'ALA': 'orange', 'ARG': 'white', 'ASN': 'white', 'ASP': 'white', 'CYS': 'orange', 'GLN': 'white', 'GLU': 'white', 'GLY': 'orange', 'HIS': 'white', 'ILE': 'orange', 'LEU': 'orange', 'LYS': 'white', 'MET': 'orange', 'PHE': 'orange', 'PRO': 'white', 'SER': 'white', 'THR': 'white', 'TRP': 'orange', 'TYR': 'orange', 'VAL': 'orange', }}
v = py3Dmol.view(query='1ubq',style={'stick':{'colorscheme':hydro}}); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
Disulfide Bonds¶
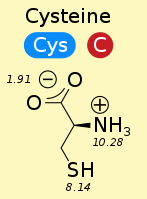
A covalent bond between the sulfurs (thiol group) of two cysteines (cross-link).
A "molecular staple"
Can act as a redox sensor - disulfid is oxidized fom and unbound is reduced.
v = py3Dmol.view(query='3rnt',style='cartoon'); v.addStyle({'resn':'CYS'},'stick'); v.zoomTo({'resn':'CYS'}); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
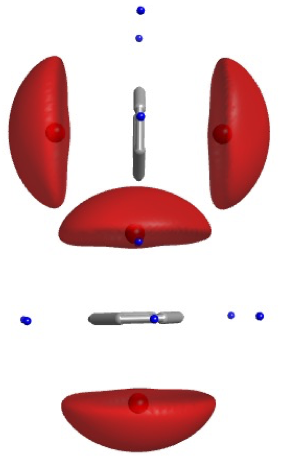
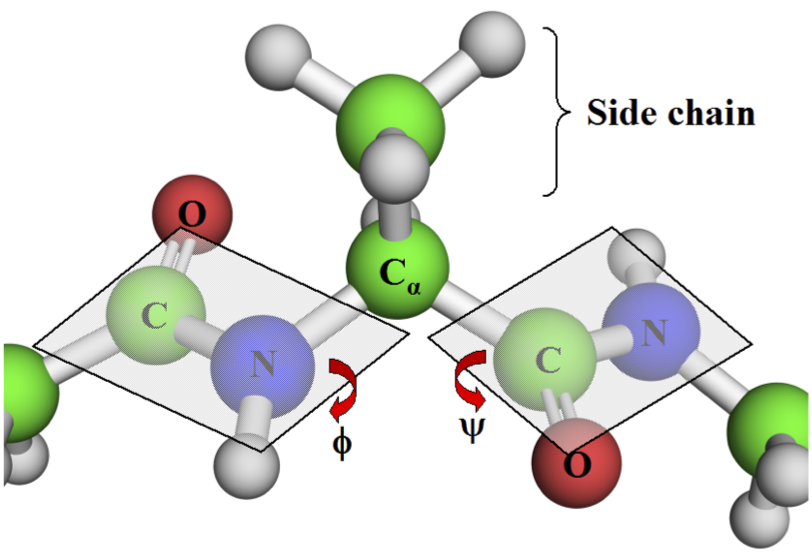
Intramolecular interactions: Dihedrals¶

- psi ($\psi$): Between backbone carbons
- phi ($\phi$): Between $C_\alpha$ and N
- omega ($\omega$): Between C (NOT $C\alpha$) and N. The peptide bond
Peptide Bond¶
Due to resonance, peptide bond has strong preference for remaining planar.
Trans is strongly preferred (except proline).
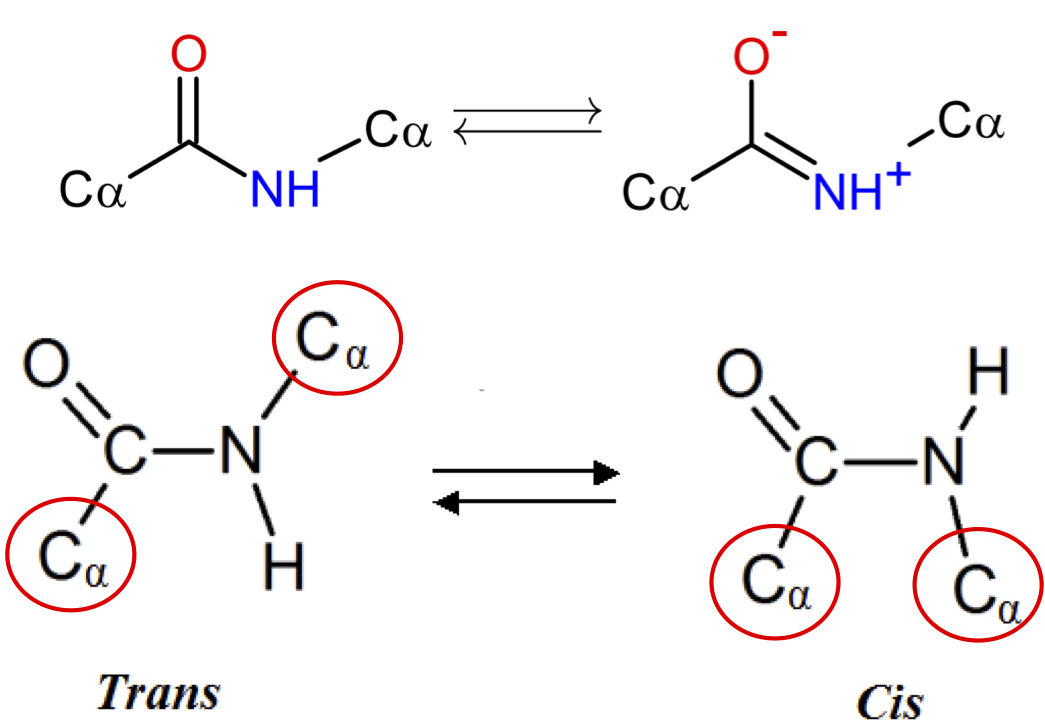
Why is trans preferred?
From Introduction to Proteins: Structure, Function, and Motion. Amit Kessel & Nir Ben-Tal
Ramachandran Plot¶
As the backbone geometry is largely determined by $\phi$ and $\psi$, can plot their propensities and observe there are clear preferences.

v = py3Dmol.view(query='3ssi',style='cartoon'); v.addStyle({'resn':'GLY'},'sphere'); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
v = py3Dmol.view(query='5cts',style='none'); v.setStyle({'resi':'5-29'},'cartoon'); v.addStyle({'resi':15},'stick'); v.zoomTo({'resi':'5-29'}); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
Protein Structure¶
Molecular interactions result in a hierarchy of of structures:
- Primary structure: The sequence
- Secondary structure: The local conformation (helix/sheet/loop)

- Tertiary structure: The complete fold of a single protein chain
- Quaternary structure: The arrangement of multiple chains
Protein Domain¶

A whole or partial peptide chain that forms an independent structural unit
- well-defined hydrophobic core (usually)
- specific function (usually)
- building block of evolution
Secondary Structure: Alpha Helices¶

- ~30% of residues in globular proteins
- 3.6 residues per turn
- $2.5\mathrm{\mathring{A}}$ radius
- N-H to C=O hydrogen bond between residues $i$ and $i+4$
- $\phi \approx -60^\circ$
- $\psi \approx -40^\circ$
Secondary Structure: Weird Helices¶
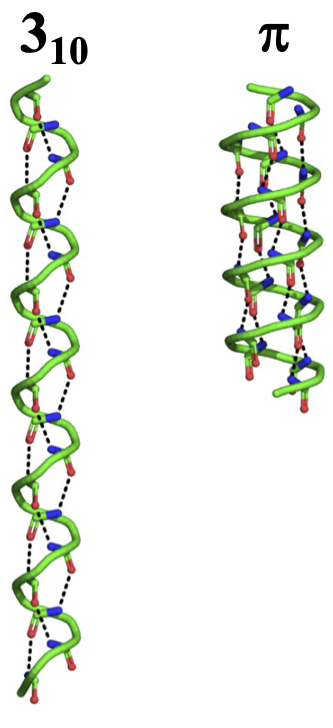
These are not common (energetically unfavorable) and are usually small and at the start/end of an alpha helix.
$\pi$ helix¶
- Less tightly wound
- H-bond between $i$ and $i+5$
- 4.4 residues/turn
- $2.8\mathrm{\mathring{A}}$ radius
$3_{10}$ helix¶
- More tightly wound
- H-bond between $i$ and $i+3$
- 3.0 residues/turn
- $1.9\mathrm{\mathring{A}}$ radius
v = py3Dmol.view(query='1cag',style={'cartoon':{'colorscheme':'chain'}}); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
v = py3Dmol.view(query='1cag',style={'stick':{'colorscheme':'chain'}}); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
Secondary Structure: Beta Strand¶
- Second most common after alpha helices (~20% of residues in globular proteins)
- Extended backbone
- Alternating side-chains
- $\phi \approx -120^\circ$
- $\psi \approx 120^\circ$
- Form sheets
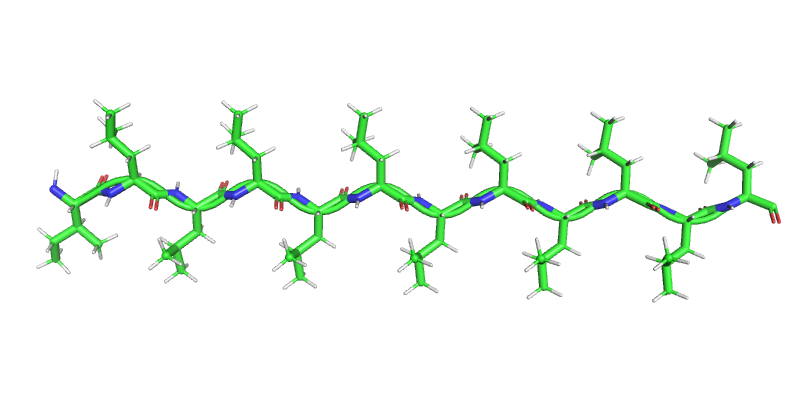
Beta Sheet: Antiparallel¶
- Strands run in diffent directions
- Unevenly spaced hydrogen bonds
- Well orient hydrogen bonds - slightly more stable
v = py3Dmol.view(query='1f94',style={'cartoon':{'color':'spectrum','arrows':True},'stick':{}}); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
Beta Sheet: Parallel¶
- Strands run in the same direction
- Evenly spaced hydrogen bonds
- Slighly less stable than anti-parallel (non-ideal h-bonds)
v = py3Dmol.view(query='1tph',style='none'); v.setStyle({'chain':'1'},{'cartoon':{'color':'spectrum','arrows':True},'stick':{'radius':0.2}}); v.zoomTo({'chain':'1'}); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
Structural Motifs¶
Specific geometric arrangements of secondary structure that occur frequently (and someone has bothered to name)
Helix-Turn-Helix Motif¶
v = py3Dmol.view(query='1DU0',style='cartoon'); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
Coiled-Coil¶
v = py3Dmol.view(query='1C1G',style={'cartoon':{'colorscheme':'chain'}}); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
Beta Barrel¶
v = py3Dmol.view(query='1BRP',style={'cartoon':{'color':'spectrum'}}); v.show()
3Dmol.js failed to load for some reason. Please check your browser console for error messages.
%%html
<div id="whatdrives" style="width: 500px"></div>
<script>
var divid = '#whatdrives';
jQuery(divid).asker({
id: divid,
question: "What do you think is more responsible for secondary structure formation?",
answers: ["Hydrogen bonding","Hydrophobicity","Both","Neither"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>